Getting Started
This tutorial walks through running your first thor simulation.
Prerequisites
- Thor built successfully (see Installation)
Your First Simulation
Thor is primarily configured via a single configuration file that we pass to thor on start-up. There's a number of compile-time choices, which we will not discuss (for now).
Note that there is no dedicated tutorial material yet. As substitute, it can be helpful to explore configurations in regression_tests/CI.
1. Create a configuration file
Create my_first_sim.yaml:
dataset_type: 'shellmodel'
driver_type: 'mcrtsimulation'
device: "cpu"
log_level: info
shellmodel:
boxsize: 3.086e21 # cm (1 kpc)
inner_radius: 0.0
outer_radius: 0.1
density: 0.078098 # cm^-3 (tau ~ 1e6)
temperature: 20000.0
density_fluff: 0.0
temperature_fluff: 0.0
outflow_velocity: 0.0
mcrtsimulation:
outputpath: output
overwrite: true
linename: Lya
nphotons_max: 10000
nphotons_step_max: 10000
nsteps_per_photon_max: 10000000
acc_scheme: Smith15
emissionmodel:
mode: "singlesource"
singlesource:
nphotons: 1000
lum_total: 1e42
position: [0.5, 0.5, 0.5]
This config has been copied and adjusted from ./regression_tests/CI/shellmodel/config.yaml.
2. Run the simulation
(Optionally) move the config file to some folder, where you want the results to be saved. Then execute:
You will need to adjust thor to the position of your compiled binary. If you followed the previous installation guide, the binary should be in ./build/src/thor.
Thor will give you some intermediate status update during the calculation. Once finished, you should find the raw photon output under ./output. For given example, you will find two subfolders: One for the initially spawned photons (input; right after emission) and those after radiative transfer (original; those escaping the simulation domain). By default, these results are in hdf5 format, and consist of one-dimensional datasets e.g.
> h5glance original/data.h5
original/data.h5
├column_density_seen [float64: 1000]
├current_cell [int64: 1000]
├direction [float64: 1000 × 3]
├dlambda [float64: 1000]
├id [int64: 1000]
├lsp_dist [float64: 1000]
├next_cell [int64: 1000]
├nscatterings [int32: 1000]
├nsteps [int64: 1000]
├origin [float64: 1000 × 3]
├position [float64: 1000 × 3]
├state [uint16: 1000]
├tau_seen [float64: 1000]
├tau_target [float64: 1000]
└weight [float64: 1000]
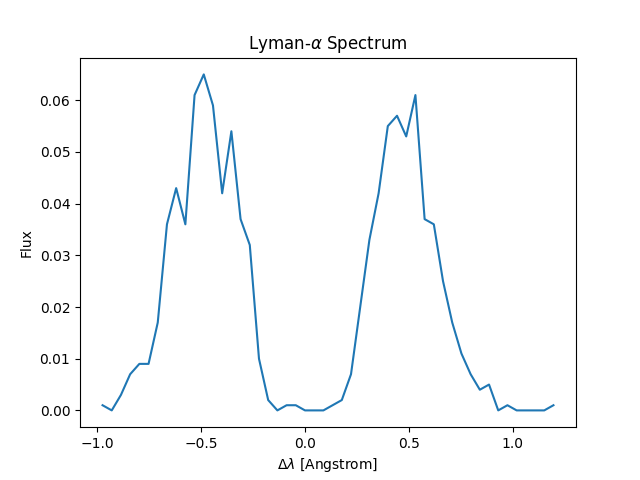
3. Visualize the spectrum
Create a simple Python script to plot the Lyman-alpha spectrum:
import h5py
import numpy as np
import matplotlib.pyplot as plt
with h5py.File("original/data.h5", "r") as f:
dlambda = f["dlambda"][:]
weight = f["weight"][:]
hist, bin_edges = np.histogram(dlambda, bins=50, weights=weight)
bin_centers = 0.5 * (bin_edges[:-1] + bin_edges[1:])
plt.plot(bin_centers, hist)
plt.xlabel(r"$\Delta\lambda$ [Angstrom]")
plt.ylabel("Flux [arbitrary]")
plt.title(r"Lyman-$\alpha$ Spectrum")
plt.tight_layout()
plt.savefig("spectrum.png")
plt.show()
Next Steps
- Configuration Reference - All available options
- Datasets - Detailed dataset documentation
- Examples - More complex setups